A wonderful little bash problem
Background
In working with fish environmental DNA, I need to have reference databases of known DNA so I can look up the species of each piece of DNA we sample. The ‘main’ reference database is GenBank. I work partially as a Data Engineer, curating our own internal database: we have some marker genes we look for, we do not expect non-Australian species in our eDNA, many public sequences are mislabeled and need to be removed, and so on. There’s an API named Entrez to talk to GenBank via a set of command-line tools called eutils. I wrote a Python script that takes a set of ‘our’ Australian taxonomy IDs and writes a bash script that calls eutils.
The problem
I changed my eutils command slightly to remove unnecessary filtering and suddenly the API returned a lot of nonsense.
See if you can spot the problem:
$ esearch -db nuccore -query “(txid29146[ORGN] OR txid107764[ORGN][…heaps more taxids] ) AND (“cytochrome c oxidase 1”[Title] OR “cytochrome oxidase subunit I”[Title] OR COI[Title] OR COXI[Title] OR COX1[Title] OR “COX 1”[Title] OR “COX I”[Title] OR CO1[Title] OR C01[Title] OR “cytochrome oxidase I”[Title] OR “cytochrome oxidase subunit I”[Title] OR “cytochrome oxidase subunit 1”[Title] OR “cytochrome oxidase 1”[Title] OR “cytochrome c oxidase subunit I”[Title] OR “cytochrome c oxidase subunit 1”[Title])” | efetch -format fasta
This command was generated by a Python script:
dl_string = dl_string.rstrip(‘OR ‘) + ‘) AND (\“cytochrome c oxidase 1\"[Title] OR \“cytochrome oxidase subunit I\"[Title] OR COI[Title] OR COXI[Title] OR COX1[Title] OR \“COX 1\"[Title] OR \“COX I\"[Title] OR CO1[Title] OR C01[Title] OR \“cytochrome oxidase I\"[Title] OR \“cytochrome oxidase subunit I\"[Title] OR \“cytochrome oxidase subunit 1\"[Title] OR \“cytochrome oxidase 1\"[Title] OR \“cytochrome c oxidase subunit I\"[Title] OR \“cytochrome c oxidase subunit 1\"[Title])\” | efetch -format fasta )’
Can you spot the error?
The solution
I had double quotes within double quotes. Bash removes those!
$ echo “hey “ho””
is evaluated to
hey ho
In my above search commands, this means that “cytochrome oxidase subunit I” as a single search-term turned into four search terms, “cytochrome”, “oxidase”, “subunit”, and “I”. All kinds of genes have “subunit” and “I” in their name! That’s why the Entrez API returned all kinds of random genes.
This is partially an issue caused by using Python to write bash scripts; I escape the double quotes via \”, but that fixes only quotes within quotes in Python, not bash. I would’ve needed to write \\” to keep the \” intact inside the bash script. A Matryoshka doll of quotes.
This is why Python packages like subprocess exist, there’s really no need to write bash scripts via Python. If I would’ve done it properly in the first place, I would have never had this problem. (The lazier solution is to replace the double quotes around the search term with single quotes - which is what I’ve done now, and must’ve removed when I updated the script.)
P.S.: Since bash removes double quotes within double quotes you get the illusion that Python-style triple quotes work:
$ echo “““hi”””
evaluates to
hi
But in reality, those double quotes just cancel each other out.
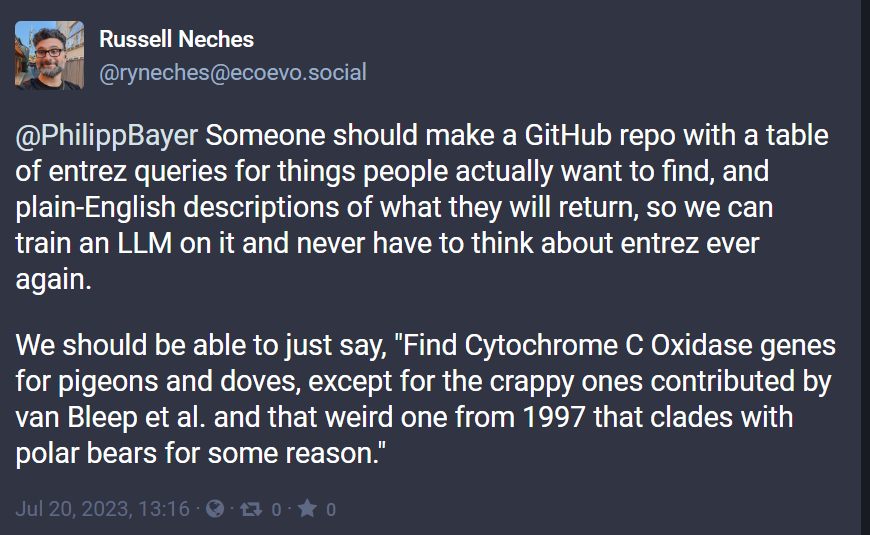
P.P.S: As Russell Neches points out on Mastodon, Entrez queries are full of little pitfalls. Time for some out-of-the-box thinking!
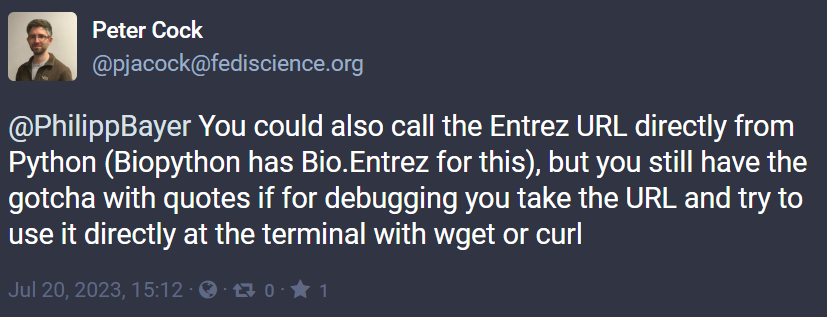
P.P.P.S: As Peter J Cock points out on Mastodon, I could’ve also just used Bio.Entrez within Biopython. I might’ve hit similar bugs once I would’ve debugged on the command line, though.