GO enrichment
Philipp Bayer
01/10/2021
Last updated: 2023-11-22
Checks: 6 1
Knit directory:
Amphibolis_Posidonia_Comparison/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of
the R Markdown file created these results, you’ll want to first commit
it to the Git repo. If you’re still working on the analysis, you can
ignore this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210414) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version c347565. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rproj.user/
Ignored: renv/library/
Ignored: renv/staging/
Unstaged changes:
Modified: analysis/GO_and_ortho.Rmd
Modified: analysis/GOenrichment.Rmd
Modified: analysis/index.Rmd
Modified: analysis/metagenome.Rmd
Modified: data/Lost_present_gene_lists/Genes_only_in_Algae.txt
Modified: data/Lost_present_gene_lists/Genes_only_in_Aquatics_and_Seagrasses.txt
Modified: data/Lost_present_gene_lists/Genes_only_in_Aquatics_and_Seagrasses_and_Terrestrials.txt
Modified: data/Lost_present_gene_lists/Genes_only_in_Duckweeds.txt
Modified: data/Lost_present_gene_lists/Genes_only_in_Seagrasses.txt
Modified: data/Lost_present_gene_lists/Genes_only_in_Terrestrials.txt
Modified: data/Lost_present_gene_lists/Genes_union_of_Seagrass_and_Aquatics_union.txt
Modified: data/arabidopsis_gene_level_comparison.xlsx
Modified: data/arabidopsis_gene_level_comparison_only_losts.xlsx
Modified: data/arabidopsis_gene_level_counts.xlsx
Modified: output/GO_results_genes_in_Algae_NOT_Duckweeds_NOT_Seagrasses_NOT_Terrestrials.csv.png
Modified: output/GO_results_genes_in_Aquatics_NOT_Seagrasses_NOT_Terrestrials.csv.png
Modified: output/GO_results_genes_in_Aquatics_and_Seagrasses_NOT_Terrestrials.csv.png
Modified: output/GO_results_genes_in_Duckweeds_NOT_Algae_NOT_Seagrasses_NOT_Terrestrials.csv.png
Modified: output/GO_results_genes_in_Seagrasses_NOT_Algae_NOT_Duckweeds_NOT_Terrestrials.csv.png
Modified: output/GO_results_genes_in_Seagrasses_NOT_Aquatics_NOT_Terrestrials.csv.png
Modified: output/GO_results_genes_in_Terrestrials_NOT_Aquatics_NOT_Seagrasses.csv.png
Modified: output/GO_results_genes_in_all_seagrasses_vs_backgroundAll_intersect.csv.png
Modified: output/GO_results_genes_in_all_seagrasses_vs_backgroundAll_union.csv.png
Modified: output/GO_results_genes_in_all_seagrasses_vs_seagrassesBackground_intersect.csv.png
Modified: output/GO_results_genes_in_all_seagrasses_vs_seagrassesBackground_union.csv.png
Modified: output/GO_results_genes_lost_A_antarctica_not_other_seagrasses.csv.png
Modified: output/GO_results_genes_lost_P_australis_not_other_seagrasses.csv.png
Modified: output/GO_results_genes_lost_Z_marina_not_other_seagrasses.csv.png
Modified: output/GO_results_genes_lost_Z_muelleri_not_other_seagrasses.csv.png
Modified: output/GO_results_genes_only_P_australis_not_other_seagrasses.csv.png
Modified: output/GO_results_genes_only_Z_marina_not_other_seagrasses.csv.png
Modified: output/GO_results_genes_only_Z_muelleri_not_other_seagrasses.csv.png
Modified: output/GO_results_genes_union_Aquatics_and_Seagrasses_NOT_Terrestrials.csv.png
Modified: output/GO_results_terrestrials_vs_union_aquatics_seagrasses.png
Modified: output/Lost_GO_terms_in_five_species.PlantSpecific.xlsx
Modified: output/Lost_GO_terms_in_five_species.xlsx
Modified: output/Seagrasses_shared_lost_genes.xlsx
Modified: output/all_GO_plots.Rdata
Modified: output/group_venn_image.Rdata
Modified: output/seagrass_venn_image.Rdata
Staged changes:
Modified: output/patchwork_seagrass_gene_loss.png
Modified: output/patchwork_terrestrials_gene_loss.png
New: output/patchwork_terrestrials_seagrasses_gene_loss.png
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/GOenrichment.Rmd) and HTML
(docs/GOenrichment.html) files. If you’ve configured a
remote Git repository (see ?wflow_git_remote), click on the
hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 0e47209 | Philipp Bayer | 2022-04-26 | Build site. |
| html | 663dcbc | Philipp Bayer | 2022-03-10 | Build site. |
| Rmd | d4cd865 | Philipp Bayer | 2022-03-10 | make the setup blocks nicer |
| html | aca07fd | Philipp Bayer | 2022-03-10 | Build site. |
| Rmd | be91c66 | Philipp Bayer | 2022-03-10 | GO-term updates! more groups! for the price of one! |
| Rmd | 2edd81b | Philipp Bayer | 2022-03-09 | add more ifles |
| Rmd | cec9544 | Philipp Bayer | 2022-03-01 | Add general descriptions |
| html | 5b15384 | Philipp Bayer | 2022-02-28 | Build site. |
| Rmd | 581bebd | Philipp Bayer | 2022-02-28 | accidentally did not display REVIGO code |
| html | 6eeb3ed | Philipp Bayer | 2022-02-25 | Add missing files |
| html | 9ca6c94 | Philipp Bayer | 2022-02-25 | Build site. |
| Rmd | 61c8f69 | Philipp Bayer | 2022-02-25 | Replace GO enrichment |
| html | 9b59467 | Philipp Bayer | 2022-02-23 | Rewrite Orthofinder analysis |
| Rmd | ab9a1f6 | Philipp Bayer | 2022-02-21 | add some more changes |
| html | 65d3fa4 | Philipp Bayer | 2021-12-20 | Build site. |
| Rmd | 21f4c6f | Philipp Bayer | 2021-12-20 | fix tables again, finally :) haha |
| html | d8ec8d3 | Philipp Bayer | 2021-12-20 | Build site. |
| Rmd | 2fb63eb | Philipp Bayer | 2021-12-20 | fix tables again, finally :) |
| html | cf2e045 | Philipp Bayer | 2021-12-20 | Build site. |
| Rmd | 280fc5d | Philipp Bayer | 2021-12-20 | fix tables again |
| html | 5356e2c | Philipp Bayer | 2021-12-20 | Build site. |
| Rmd | 9f4be6f | Philipp Bayer | 2021-12-20 | fix tables |
| html | 7816b8c | Philipp Bayer | 2021-12-20 | Build site. |
| Rmd | 73bffa4 | Philipp Bayer | 2021-12-20 | add tables |
| html | c1288a4 | Philipp Bayer | 2021-12-20 | Build site. |
| Rmd | 2691cad | Philipp Bayer | 2021-12-20 | add tables |
| html | 292db59 | Philipp Bayer | 2021-10-18 | Build site. |
| Rmd | 35aa72f | Philipp Bayer | 2021-10-18 | wflow_publish(c("analysis/GOenrichment.Rmd", "analysis/plotGenes.Rmd", |
| Rmd | 7e370e9 | Philipp Bayer | 2021-10-07 | Updated GOenrichment |
| Rmd | 95586f9 | Philipp Bayer | 2021-10-07 | Add missing data |
| html | 95586f9 | Philipp Bayer | 2021-10-07 | Add missing data |
| html | c0db4a5 | Philipp Bayer | 2021-10-07 | Build site. |
| Rmd | 08b28ea | Philipp Bayer | 2021-10-07 | wflow_publish(files = c("analysis/*")) |
library(topGO)
library(tidyverse)
library(rvest)
library(rrvgo)
library(wesanderson)
library(httr)
library(stringi)
library(ggrepel)
library(eulerr)
library(UpSetR)
library(rrvgo)
library(kableExtra)
library(patchwork)
knitr::opts_knit$set(root.dir = rprojroot::find_rstudio_root_file())Lost in seagrasses
The GO enrichment does not work well on my laptop so I’m setting this to eval=FALSE and run it on a remote server.
This document takes the output of Orthofinder
# give properly formatted background in format: GO:0005838 GSBRNA2T00088508001;GSBRNA2T00088313001;GSBRNA2T00035842001
#annAT <- readMappings('BACKGROUND.txt.gz', sep="\t", IDsep=";")
#save(annAT, file='annAtObject.RData')
load('annAtObject.RData')
allgenes <- unique(unlist(annAT))
compare <- function(genelistfile, outname, allgenes, annAT) {
# give file with your genes of interest, one gene_id per line
mygenes <-scan(genelistfile ,what="")
geneList <- factor(as.integer(allgenes %in% mygenes))
names(geneList) <- allgenes
GOdata <-new ("topGOdata", ontology = 'BP', allGenes = geneList, nodeSize = 5, annot=annFUN.GO2genes, GO2genes=annAT)
# using ClassicCount:
#test.stat <-new ("classicCount", testStatistic = GOFisherTest, name = "Fisher Test")
#resultsFisherC <-getSigGroups (GOdata, test.stat)
# using weight01:
weight01.fisher <- runTest(GOdata, statistic = "fisher")
# using ClassicCount:
# allRes <- GenTable(GOdata, classicFisher= resultsFisherC, topNodes = 30)
# using weight01:
allRes <- GenTable(GOdata, classicFisher=weight01.fisher,topNodes=30)#,topNodes=100)
names(allRes)[length(allRes)] <- "p.value"
p_values <- score(weight01.fisher)
adjusted_p <- p.adjust(p_values)
adjusted_p[adjusted_p < 0.05] %>% enframe() %>% write_csv('data/' + outname)
}Comparing the four big groups
# all genes shared - this runs out of memory
#compare('Lost_present_gene_lists/Genes_only_in_Aquatics_and_Seagrasses_and_Terrestrials.txt', 'output/GO_results_genes_in_Aquatics_and_Seagrasses_and_Terrestrials.csv', allgenes, annAT)
# Genes in aquatics and seagrasses - IMPORTANT This is NOT the union - this is the intersection!
# Union is below
compare('Lost_present_gene_lists/Genes_only_in_Aquatics_and_Seagrasses.txt', 'output/GO_results_genes_in_Aquatics_and_Seagrasses_NOT_Terrestrials.csv', allgenes, annAT)
# Genes only in algae
compare('Lost_present_gene_lists/Genes_only_in_Algae.txt', 'output/GO_results_genes_in_Algae_NOT_Duckweeds_NOT_Seagrasses_NOT_Terrestrials.csv', allgenes, annAT)
# Genes only in seagrasses
compare('Lost_present_gene_lists/Genes_only_in_Seagrasses.txt', 'output/GO_results_genes_in_Seagrasses_NOT_Algae_NOT_Duckweeds_NOT_Terrestrials.csv', allgenes, annAT)
# Genes only in duckweeds
compare('Lost_present_gene_lists/Genes_only_in_Duckweeds.txt', 'output/GO_results_genes_in_Duckweeds_NOT_Algae_NOT_Seagrasses_NOT_Terrestrials.csv', allgenes, annAT)
# Genes only in terrestrials
compare('Lost_present_gene_lists/Genes_only_in_Terrestrials.txt', 'output/GO_results_genes_in_Terrestrials_NOT_Aquatics_NOT_Seagrasses.csv', allgenes, annAT)
# Genes union of Seagrass and Aquatics
compare('Lost_present_gene_lists/Genes_union_of_Seagrass_and_Aquatics_union.txt', 'output/GO_results_genes_union_Aquatics_and_Seagrasses_NOT_Terrestrials.csv', allgenes, annAT)
# Genes only in seagrasses- seagrass-specific
compare('Lost_present_gene_lists/Genes_in_all_seagrasses_intersect.txt', 'output/GO_results_genes_in_all_seagrasses_vs_backgroundAll_intersect.csv', allgenes, annAT)
compare('Lost_present_gene_lists/Genes_in_all_seagrasses_union.txt', 'output/GO_results_genes_in_all_seagrasses_vs_backgroundAll_union.csv', allgenes, annAT)Seagrass only comparisons
Now we compare seagrasses within each other.
For the seagrass-only comparisons, I’m using a Seagrass-only background as that makes more biological sense to me.s
# give properly formatted background in format: GO:0005838 GSBRNA2T00088508001;GSBRNA2T00088313001;GSBRNA2T00035842001
#sannAT <- readMappings('SEAGRASSBACKGROUND.txt', sep="\t", IDsep=";")
#save(sannAT, file='sannAtObject.RData')
load('sannAtObject.RData')
sallgenes <- unique(unlist(sannAT))lost in seagrasses
compare('Lost_present_gene_lists/Genes_lost_in_A_antarctica_not_other_seagrasses.txt', 'output/GO_results_genes_lost_A_antarctica_not_other_seagrasses.csv', sallgenes, sannAT)
compare('Lost_present_gene_lists/Genes_lost_in_Z_marina_not_other_seagrasses.txt', 'output/GO_results_genes_lost_Z_marina_not_other_seagrasses.csv', sallgenes, sannAT)
compare('Lost_present_gene_lists/Genes_lost_in_P_australis_not_other_seagrasses.txt', 'output/GO_results_genes_lost_P_australis_not_other_seagrasses.csv', sallgenes, sannAT)
compare('Lost_present_gene_lists/Genes_lost_in_Z_muelleri_not_other_seagrasses.txt', 'output/GO_results_genes_lost_Z_muelleri_not_other_seagrasses.csv', sallgenes, sannAT)Present in seagrasses
compare('Lost_present_gene_lists/Genes_in_all_seagrasses_intersect.txt', 'output/GO_results_genes_in_all_seagrasses_vs_seagrassesBackground_intersect.csv', sallgenes, sannAT)
compare('Lost_present_gene_lists/Genes_in_all_seagrasses_union.txt', 'output/GO_results_genes_in_all_seagrasses_vs_seagrassesBackground_union.csv', sallgenes, sannAT)
compare('Lost_present_gene_lists/Genes_only_in_Z_marina_not_other_seagrasses.txt', 'output/GO_results_genes_only_Z_marina_not_other_seagrasses.csv', sallgenes, sannAT)
compare('Lost_present_gene_lists/Genes_only_in_Z_muelleri_not_other_seagrasses.txt', 'output/GO_results_genes_only_Z_muelleri_not_other_seagrasses.csv', sallgenes, sannAT)
compare('Lost_present_gene_lists/Genes_only_in_P_australis_not_other_seagrasses.txt', 'output/GO_results_genes_only_P_australis_not_other_seagrasses.csv', sallgenes, sannAT)
compare('Lost_present_gene_lists/Genes_only_in_A_antarctica_not_other_seagrasses.txt', 'output/GO_results_genes_only_A_antarctica_not_other_seagrasses.csv', sallgenes, sannAT)Alright now we have all these different GO terms in all these files - we can send them to revigo for visualiation and some deduplication!
Revigo
This code is based on http://revigo.irb.hr/CodeExamples/revigo.R.txt
results_list <- list()
for (f in list.files('./output/', pattern='*.csv')){
if(str_detect(f, '.png')) {
next
}
filename <- paste('./output/', f, sep='')
go_and_pvalues <- readChar(filename, file.info(filename)$size)
go_and_pvalues <- gsub(',', ' ', go_and_pvalues)
temp <- read_delim(go_and_pvalues,show_col_types = FALSE)
if(nrow(temp) < 2) {next}
simMatrix <- calculateSimMatrix(temp$name,
orgdb="org.At.tair.db",
ont="BP",
method="Rel")
tmp <- temp$value
names(tmp) <- temp$name
reducedTerms <- reduceSimMatrix(simMatrix,
tmp,
threshold=0.7,
orgdb="org.At.tair.db")
results_list[[f]] <- list('simMatrix'=simMatrix, 'reducedTerms'=reducedTerms)
#httr::POST(
# url = "http://revigo.irb.hr/StartJob",
# body = list(
# cutoff = "0.7",
# valueType = "pvalue",
# # speciesTaxon = "4577", # zea mays
# #speciesTaxon = '39947', # japonica
# speciesTaxon = '3702', # arabidopsis
# measure = "SIMREL",
# goList = go_and_pvalues
# ),
# # application/x-www-form-urlencoded
# encode = "form"
#) -> res
#
# dat <- httr::content(res, encoding = "UTF-8")
#
# if(typeof(dat)!="list")
# {
# jobid <- jsonlite::fromJSON(dat,bigint_as_char=TRUE)$jobid
# } else {
# jobid <- dat$jobid
# }
# # Check job status
# running <- "1"
# while (running != "0" ) {
# httr::GET(
# url = "http://revigo.irb.hr/QueryJob",
# query = list( jobid = jobid, type="jstatus" )
# ) -> res2
# dat2 <- httr::content(res2, encoding = "UTF-8")
# # fix when httr::content automatically converts json to list
# if(typeof(dat2)!="list")
# {
# running <- jsonlite::fromJSON(dat2)$running
# } else {
# running <- dat2$running
# }
# Sys.sleep(1)
# }
#
# # Fetch results
# httr::GET(
# url = "http://revigo.irb.hr/QueryJob",
# query = list(
# jobid = jobid,
# namespace = "1",
# type = "table"
# )
# ) -> res3
# dat3 <- httr::content(res3, encoding = "UTF-8")
#
# dat3 <- stri_replace_all_fixed(dat3, "\r", "")
# # Now we have a csv table in a string!
#
# # read_csv does not like the ', ', it wants ','
# dat <- read_tsv(gsub(', ', ',', dat3), show_col_types = FALSE)
#
# # do we even have results?
# if(nrow(dat) == 0){next}
# results_list[[f]] <- dat
#
}preparing gene to GO mapping data...preparing IC data...preparing gene to GO mapping data...preparing IC data...preparing gene to GO mapping data...preparing IC data...preparing gene to GO mapping data...preparing IC data...preparing gene to GO mapping data...preparing IC data...preparing gene to GO mapping data...preparing IC data...preparing gene to GO mapping data...preparing IC data...preparing gene to GO mapping data...preparing IC data...preparing gene to GO mapping data...preparing IC data...preparing gene to GO mapping data...preparing IC data...preparing gene to GO mapping data...preparing IC data...preparing gene to GO mapping data...preparing IC data...preparing gene to GO mapping data...preparing IC data...preparing gene to GO mapping data...preparing IC data...preparing gene to GO mapping data...preparing IC data...preparing gene to GO mapping data...preparing IC data...preparing gene to GO mapping data...preparing IC data...preparing gene to GO mapping data...preparing IC data...OK we have a list with all results in a big list. Now we can plot!
# lots of warnings and messages here so I'm hiding these
plot_list <- list()
for (f in names(results_list)) {
tmp <- results_list[[f]]
simMatrix <- tmp[['simMatrix']]
reducedTerms <- tmp[['reducedTerms']]
if(nrow(reducedTerms) <= 2) {next}
plot_list[[f]] <- scatterPlot(simMatrix, reducedTerms, labelSize=3)
# old REVIGO code
# dat <- results_list[[f]]
# if(nrow(dat) == 0) {next}
#
# names(dat) <- c("term_ID","description","frequency", 'plot_X', 'plot_Y', 'log_size', 'value', 'uniqueness')#, 'dispensability', 'eliminated', 'representative')
# one.data <- dat
# #if (one.data[1,] == '<html>') {next} # some datasets result in error
# #one.data <- one.data [(one.data$plot_X != "null" & one.data$plot_Y != "null") & (! is.na(one.data$frequency)) & (one.data$value != 'null'), ];
# #one.data <- suppressMessages(type_convert(one.data)) # guess data types, but also: shush
# top_ten_values <- one.data %>% arrange(value) %>% head(8) %>% pull(term_ID)
#
# one.data <- one.data %>% mutate(description2 = case_when(
# term_ID %in% top_ten_values ~ str_wrap(description, width=10),
# TRUE ~ ''))
# p1 <-
# ggplot(data = one.data, aes(plot_X, plot_Y, label = description2)) +
# geom_point(aes(colour = value, size = log_size), alpha = I(0.6)) +
# scale_size_area() +
# scale_colour_gradientn(colours = rev(wes_palette("Zissou1", 100, type = "continuous")))+ #,
# #limits = c(min(one.data$value), 0)) +
# geom_point(
# aes(plot_X, plot_Y, size = log_size),
# shape = 21,
# fill = "transparent",
# colour = I (alpha ("black", 0.6))
# ) +
# scale_size_area() +
# geom_label_repel(max.overlaps=15,
# point.padding = 0,
# min.segment.length = 0, # draw all line segments
# aes(point.size=log_size), alpha=0.9) +
# theme_minimal() +
# labs(color = 'log(p-value)',
# size = 'Frequency') +
# theme(
# axis.text.x = element_blank(),
# axis.ticks.x = element_blank(),
# axis.text.y = element_blank(),
# axis.ticks.y = element_blank(),
# axis.title.x = element_blank(),
# axis.title.y = element_blank()
# ) +
# NULL
# #ggtitle(f)
#
# plot_list[[f]] <- p1
#
}Making Supplementary Tables for the REVIGO results
Let’s also pull these terms out as tables. Value is the
unadjusted p-value (-3.7 = 10 * -3.7)
for( i in names(results_list)) {
print(i)
#print(results_list[[i]] %>% dplyr::select(TermID, Name, Value) %>% arrange(Value) %>% kbl() %>% kable_styling())
print(results_list[[i]]$reducedTerms %>% dplyr::select(go, parent, score) %>% arrange(score) %>% kbl() %>% kable_styling())
cat("\n")
}| go | parent | score | |
|---|---|---|---|
| GO:0001666 | GO:0001666 | GO:0009416 | 0.0000000 |
| GO:0006869 | GO:0006869 | GO:0006606 | 0.0000000 |
| GO:0009753 | GO:0009753 | GO:0001101 | 0.0000000 |
| GO:0009416 | GO:0009416 | GO:0009416 | 0.0000000 |
| GO:0009723 | GO:0009723 | GO:0001101 | 0.0000000 |
| GO:0006396 | GO:0006396 | GO:0006396 | 0.0000000 |
| GO:0050821 | GO:0050821 | GO:0050821 | 0.0000000 |
| GO:0042254 | GO:0042254 | GO:0090143 | 0.0000000 |
| GO:0006606 | GO:0006606 | GO:0006606 | 0.0000000 |
| GO:0009093 | GO:0009093 | GO:0042759 | 0.0000001 |
| GO:0090143 | GO:0090143 | GO:0090143 | 0.0000003 |
| GO:0042814 | GO:0042814 | GO:0042814 | 0.0000012 |
| GO:0035556 | GO:0035556 | GO:0035556 | 0.0000257 |
| GO:0006636 | GO:0006636 | GO:0042759 | 0.0037737 |
| GO:0048441 | GO:0048441 | GO:0048441 | 0.0130933 |
| GO:0042759 | GO:0042759 | GO:0042759 | 0.0151474 |
| GO:0001101 | GO:0001101 | GO:0001101 | 0.0207153 |
| go | parent | score | |
|---|---|---|---|
| GO:0042908 | GO:0042908 | GO:0060919 | 0.0000237 |
| GO:0060416 | GO:0060416 | GO:0071359 | 0.0001510 |
| GO:0060919 | GO:0060919 | GO:0060919 | 0.0002064 |
| GO:0008283 | GO:0008283 | GO:0008283 | 0.0010484 |
| GO:0031338 | GO:0031338 | GO:0007033 | 0.0014469 |
| GO:0040008 | GO:0040008 | GO:0040008 | 0.0061495 |
| GO:0009827 | GO:0009827 | GO:0007033 | 0.0082377 |
| GO:0010256 | GO:0010256 | GO:0007033 | 0.0087504 |
| GO:0071359 | GO:0071359 | GO:0071359 | 0.0095624 |
| GO:0040029 | GO:0040029 | GO:0007033 | 0.0112609 |
| GO:0048481 | GO:0048481 | GO:0060321 | 0.0115297 |
| GO:0007164 | GO:0007164 | GO:0007164 | 0.0127805 |
| GO:0034644 | GO:0034644 | GO:0071480 | 0.0154031 |
| GO:0060321 | GO:0060321 | GO:0060321 | 0.0208377 |
| GO:0007033 | GO:0007033 | GO:0007033 | 0.0217608 |
| GO:0071480 | GO:0071480 | GO:0071480 | 0.0291314 |
| GO:0000724 | GO:0000724 | GO:0000724 | 0.0420358 |
| GO:0046854 | GO:0046854 | GO:0046854 | 0.0485418 |
| go | parent | score | |
|---|---|---|---|
| GO:0006278 | GO:0006278 | GO:0006278 | 0.0000000 |
| GO:0032197 | GO:0032197 | GO:0032197 | 0.0000000 |
| GO:0090501 | GO:0090501 | GO:0090501 | 0.0000000 |
| GO:0006508 | GO:0006508 | GO:0006508 | 0.0000000 |
| GO:0030837 | GO:0030837 | GO:0007033 | 0.0005896 |
| GO:0009660 | GO:0009660 | GO:0007033 | 0.0007983 |
| GO:0010098 | GO:0010098 | GO:0010098 | 0.0009919 |
| GO:0010726 | GO:0010726 | GO:0010726 | 0.0014418 |
| GO:0040029 | GO:0040029 | GO:0007033 | 0.0037805 |
| GO:0010090 | GO:0010090 | GO:0010090 | 0.0045088 |
| GO:0042908 | GO:0042908 | GO:0006887 | 0.0066418 |
| GO:0006887 | GO:0006887 | GO:0006887 | 0.0092008 |
| GO:0071359 | GO:0071359 | GO:0060416 | 0.0100883 |
| GO:1904580 | GO:1904580 | GO:1904580 | 0.0108382 |
| GO:0051214 | GO:0051214 | GO:0051214 | 0.0133473 |
| GO:0060416 | GO:0060416 | GO:0060416 | 0.0166522 |
| GO:0007033 | GO:0007033 | GO:0007033 | 0.0347384 |
| go | parent | score | |
|---|---|---|---|
| GO:0010628 | GO:0010628 | GO:0006355 | 0.0000030 |
| GO:0009651 | GO:0009651 | GO:0010048 | 0.0000187 |
| GO:0009809 | GO:0009809 | GO:0009407 | 0.0002305 |
| GO:0008283 | GO:0008283 | GO:0008283 | 0.0002999 |
| GO:0009734 | GO:0009734 | GO:2000031 | 0.0009308 |
| GO:0009407 | GO:0009407 | GO:0009407 | 0.0015592 |
| GO:2000031 | GO:2000031 | GO:2000031 | 0.0036044 |
| GO:0009637 | GO:0009637 | GO:0010048 | 0.0047664 |
| GO:0007131 | GO:0007131 | GO:0031338 | 0.0051332 |
| GO:0031338 | GO:0031338 | GO:0031338 | 0.0107653 |
| GO:0048527 | GO:0048527 | GO:0048366 | 0.0177476 |
| GO:0006355 | GO:0006355 | GO:0006355 | 0.0261990 |
| GO:0048366 | GO:0048366 | GO:0048366 | 0.0284372 |
| GO:0010048 | GO:0010048 | GO:0010048 | 0.0386478 |
| GO:0071577 | GO:0071577 | GO:0071577 | 0.0449773 |
| go | parent | score | |
|---|---|---|---|
| GO:0006518 | GO:0006518 | GO:0006508 | 1.70e-06 |
| GO:0006508 | GO:0006508 | GO:0006508 | 4.94e-05 |
| go | parent | score | |
|---|---|---|---|
| GO:0051603 | GO:0051603 | GO:0006624 | 0.0000000 |
| GO:0071482 | GO:0071482 | GO:0070914 | 0.0000870 |
| GO:0080156 | GO:0080156 | GO:0080156 | 0.0001054 |
| GO:0046785 | GO:0046785 | GO:1990019 | 0.0001205 |
| GO:0006624 | GO:0006624 | GO:0006624 | 0.0006388 |
| GO:0006597 | GO:0006597 | GO:0006597 | 0.0006766 |
| GO:1990019 | GO:1990019 | GO:1990019 | 0.0027448 |
| GO:0030174 | GO:0030174 | GO:0030174 | 0.0034408 |
| GO:0042372 | GO:0042372 | GO:0009110 | 0.0034408 |
| GO:0009110 | GO:0009110 | GO:0009110 | 0.0083491 |
| GO:0046900 | GO:0046900 | GO:0046900 | 0.0171285 |
| GO:0070914 | GO:0070914 | GO:0070914 | 0.0171576 |
| GO:0010157 | GO:0010157 | GO:0010157 | 0.0359184 |
| go | parent | score | |
|---|---|---|---|
| GO:0005983 | GO:0005983 | GO:0030174 | 0.0087939 |
| GO:0034721 | GO:0034721 | GO:0034721 | 0.0130501 |
| GO:0046686 | GO:0046686 | GO:0046686 | 0.0175974 |
| GO:0071333 | GO:0071333 | GO:0071333 | 0.0261374 |
| GO:0030174 | GO:0030174 | GO:0030174 | 0.0492303 |
| go | parent | score | |
|---|---|---|---|
| GO:0051603 | GO:0051603 | GO:0031146 | 0.0000000 |
| GO:0046785 | GO:0046785 | GO:1990019 | 0.0000000 |
| GO:0006624 | GO:0006624 | GO:0031146 | 0.0000000 |
| GO:1990019 | GO:1990019 | GO:1990019 | 0.0000000 |
| GO:0080156 | GO:0080156 | GO:0080156 | 0.0000000 |
| GO:0010157 | GO:0010157 | GO:0010157 | 0.0000031 |
| GO:0048281 | GO:0048281 | GO:0010090 | 0.0000258 |
| GO:0032989 | GO:0032989 | GO:0010090 | 0.0000310 |
| GO:0090156 | GO:0090156 | GO:0090156 | 0.0000320 |
| GO:0010090 | GO:0010090 | GO:0010090 | 0.0000799 |
| GO:1901332 | GO:1901332 | GO:0090709 | 0.0014215 |
| GO:0006278 | GO:0006278 | GO:0006278 | 0.0017328 |
| GO:0032197 | GO:0032197 | GO:0032197 | 0.0019547 |
| GO:0010286 | GO:0010286 | GO:0009408 | 0.0096276 |
| GO:0090709 | GO:0090709 | GO:0090709 | 0.0138871 |
| GO:0006537 | GO:0006537 | GO:0006537 | 0.0152019 |
| GO:0090305 | GO:0090305 | GO:0090305 | 0.0152030 |
| GO:0048194 | GO:0048194 | GO:0015770 | 0.0167839 |
| GO:0009408 | GO:0009408 | GO:0009408 | 0.0283024 |
| GO:0045332 | GO:0045332 | GO:0015770 | 0.0284194 |
| GO:0015770 | GO:0015770 | GO:0015770 | 0.0479753 |
| GO:0031146 | GO:0031146 | GO:0031146 | 0.0498006 |
| go | parent | score | |
|---|---|---|---|
| GO:0010222 | GO:0010222 | GO:0010588 | 0.0000090 |
| GO:0040007 | GO:0040007 | GO:0040007 | 0.0000423 |
| GO:2000114 | GO:2000114 | GO:2000114 | 0.0001391 |
| GO:0010588 | GO:0010588 | GO:0010588 | 0.0003670 |
| GO:0043007 | GO:0043007 | GO:0007030 | 0.0004800 |
| GO:0080119 | GO:0080119 | GO:0007030 | 0.0071445 |
| GO:0001932 | GO:0001932 | GO:0001932 | 0.0073060 |
| GO:0007030 | GO:0007030 | GO:0007030 | 0.0158798 |
| GO:0034260 | GO:0034260 | GO:0043086 | 0.0160997 |
| GO:0046459 | GO:0046459 | GO:0046459 | 0.0181702 |
| GO:0043086 | GO:0043086 | GO:0043086 | 0.0434986 |
| go | parent | score | |
|---|---|---|---|
| GO:0000398 | GO:0000398 | GO:0000398 | 0.0000558 |
| GO:0006886 | GO:0006886 | GO:0006886 | 0.0113425 |
| GO:0006334 | GO:0006334 | GO:0006334 | 0.0360022 |
Let’s have a look at all of these plots. Manually zooming in leads to ggrepel reloading labels, so on the small scale a lot of these plots don’t have labels.
for(i in names(plot_list)) {
file_name = paste('output/', i, '.png', sep='')
cowplot::save_plot(file_name, plot_list[[i]], base_height=6)
}Warning: ggrepel: 17 unlabeled data points (too many overlaps). Consider
increasing max.overlapsWarning: ggrepel: 1 unlabeled data points (too many overlaps). Consider
increasing max.overlapsWarning: ggrepel: 6 unlabeled data points (too many overlaps). Consider
increasing max.overlapsWarning: ggrepel: 8 unlabeled data points (too many overlaps). Consider
increasing max.overlapsLet me also try and make some patchworks.
# For some reason with this plot the legends aren't correctly collected by patchwork,
# so I need to manually turn one off
patchy <- (plot_list[["GO_results_genes_in_Terrestrials_NOT_Aquatics_NOT_Seagrasses.csv"]] + theme(legend.position = "none")) /
plot_list[["GO_results_genes_union_Aquatics_and_Seagrasses_NOT_Terrestrials.csv"]] + plot_layout(guides = 'collect') + plot_annotation(tag_levels = 'A')
patchyWarning: ggrepel: 39 unlabeled data points (too many overlaps). Consider
increasing max.overlaps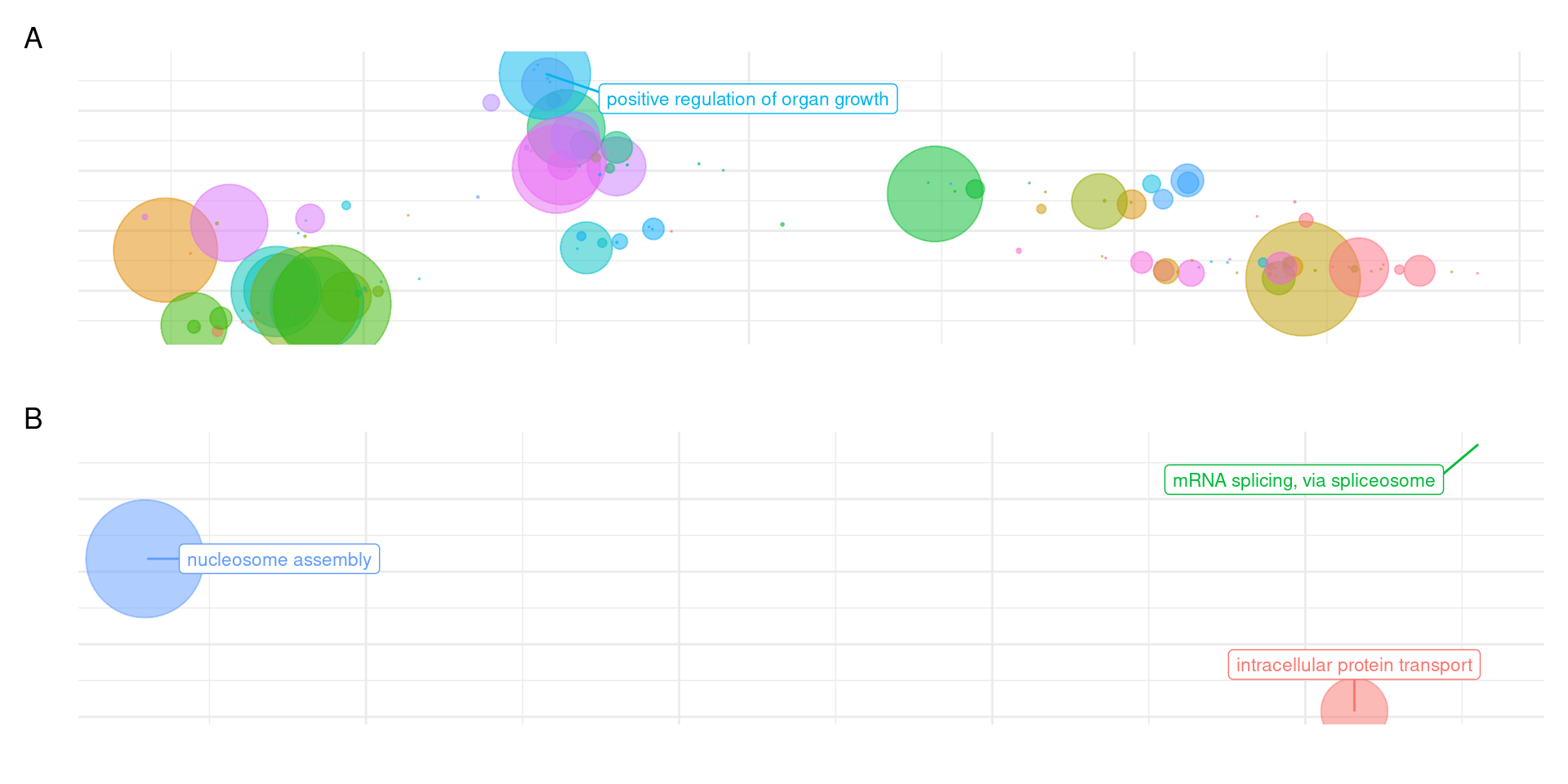
save(plot_list, file = 'output/all_GO_plots.Rdata') # save so I can put them together later
cowplot::save_plot('output/GO_results_terrestrials_vs_union_aquatics_seagrasses.png', patchy, base_height=7)Warning: ggrepel: 32 unlabeled data points (too many overlaps). Consider
increasing max.overlapsOK I’ll better make this figure for the seagrasses alone, it’s quite messy this way.
Seagrass-specific loss
Let’s check which GO terms overlap between the 4 seagrasses!
How many shared lost GO-terms are there?
a <- list(`P. australis` = results_list$GO_results_genes_lost_P_australis_not_other_seagrasses.csv$reducedTerms$term,
`A. antarctica` = results_list$GO_results_genes_lost_A_antarctica_not_other_seagrasses.csv$reducedTerms$term,
`Z. marina` = results_list$GO_results_genes_lost_Z_marina_not_other_seagrasses.csv$reducedTerms$term,
`Z. muelleri` = results_list$GO_results_genes_lost_Z_muelleri_not_other_seagrasses.csv$reducedTerms$term)
a_go_ids <- list(`P. australis` = results_list$GO_results_genes_lost_P_australis_not_other_seagrasses.csv$reducedTerms$go,
`A. antarctica` = results_list$GO_results_genes_lost_A_antarctica_not_other_seagrasses.csv$reducedTerms$go,
`Z. marina` = results_list$GO_results_genes_lost_Z_marina_not_other_seagrasses.csv$reducedTerms$go,
`Z. muelleri` = results_list$GO_results_genes_lost_Z_muelleri_not_other_seagrasses.csv$reducedTerms$go)plot(venn(a),
quantities = TRUE,
fill = rev(wes_palette("Zissou1", 15, type = 'continuous')),
alpha = 0.5,
labels = list(font = 4))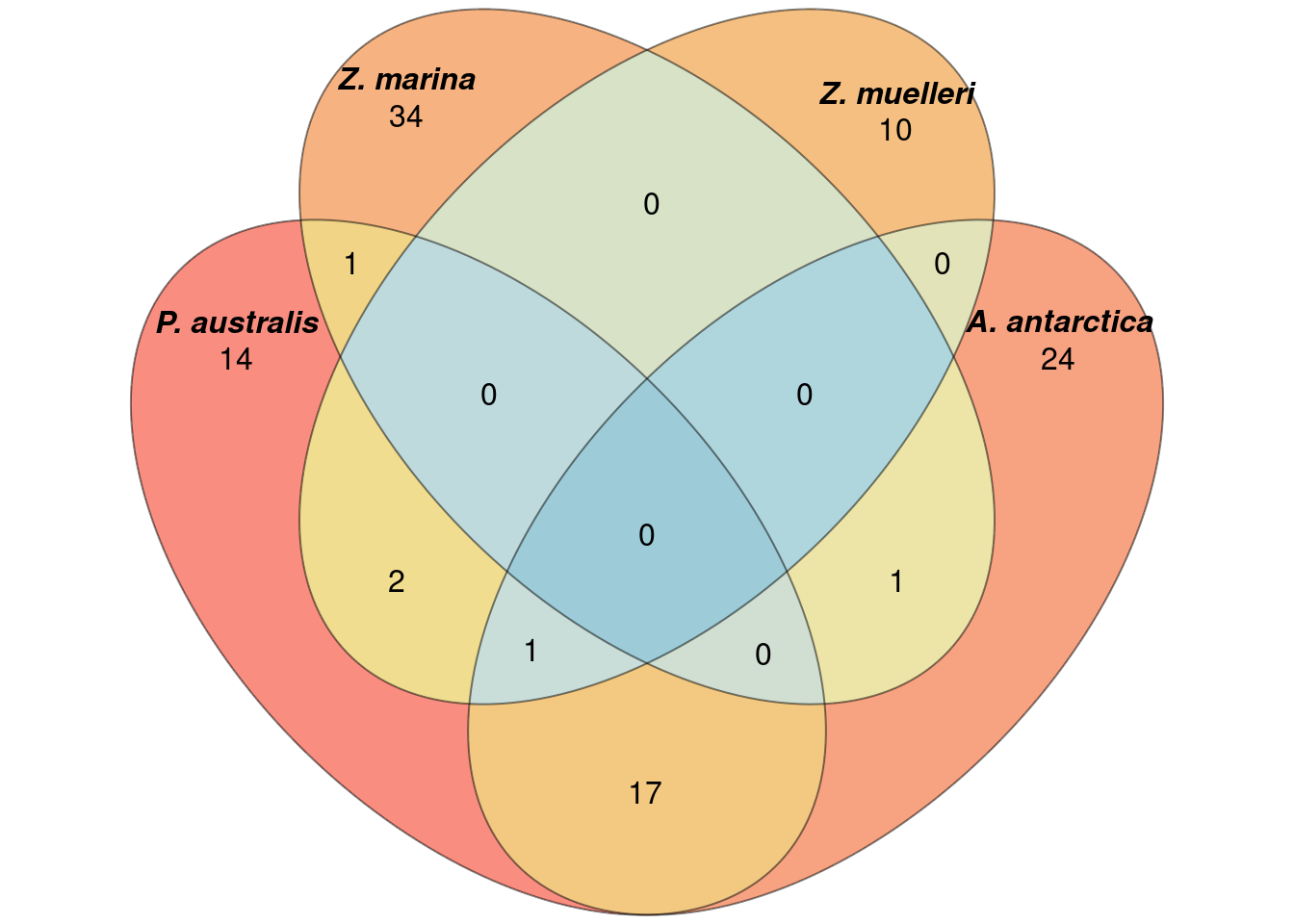
| Version | Author | Date |
|---|---|---|
| 9ca6c94 | Philipp Bayer | 2022-02-25 |
upset(fromList(a), order.by='freq', )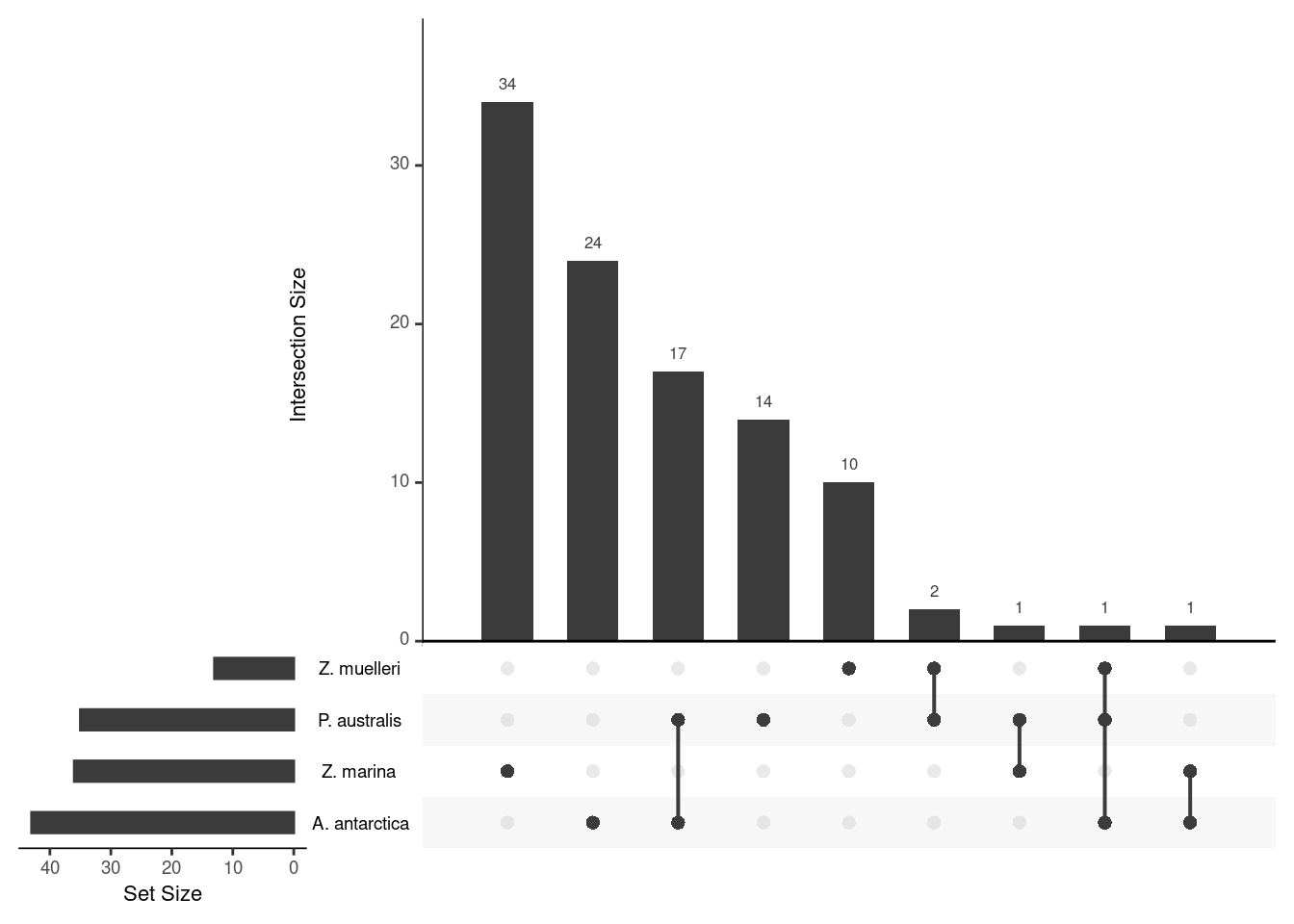
a_no_ara <- list(`P. australis` = results_list$missing_posi_vs_all_GO.txt$reducedTerms$term,
`A. antarctica` = results_list$missing_amphi_vs_all_GO.txt$reducedTerms$term,
`Z. marina` = results_list$missing_zmar_vs_all_GO.txt$reducedTerms$term,
`Z. muelleri` = results_list$missing_zmuel_vs_all_GO.txt$reducedTerms$term)What are the shared GO-terms in seagrasses, WITHOUT the Ara loss??
Reduce(union, a) %>% enframe() %>% writexl::write_xlsx('./output/Seagrasses_shared_lost_genes.xlsx')What if we remove Posidonia?
b <- list(`A. antarctica` = results_list$missing_amphi_vs_all_GO.txt$Name,
`Z. marina` = results_list$missing_zmar_vs_all_GO.txt$Name,
`Z. muelleri` = results_list$missing_zmuel_vs_all_GO.txt$Name)
intersections <- Reduce(intersect, b)
intersections[grepl('ethylene', intersections)]NULLOK we need a big list of all GO-terms here - which GO-term is lost in which species. That will be a supplementary table.
all_species <- c("P. australis","A. antarctica","Z. marina","Z. muelleri", 'A. thaliana')
all_go_terms <- Reduce(union, a)
all_go_ids <- Reduce(union, a_go_ids)
results_d <- data.frame('GOID' = character(),
'GO' = character(),
'P. australis' = character(),
'A. antarctica' = character(),
'Z. marina' = character(),
'Z. muelleri' = character(),
'A. thaliana' = character())
for (index in seq_along(all_go_terms)){
go <- all_go_terms[index]
go_id <- all_go_ids[index]
specs <- c()
for (species in names(a)) {
if ( length(a[[species]][grep(paste('^', go, '$', sep=''), a[[species]])]) > 0 ) {
specs <- c(specs, species)
}
}
results_d[index,] <- c(go_id, go, gsub('FALSE', 'Present', gsub('TRUE', 'Lost', all_species %in% specs)))
}writexl::write_xlsx(results_d, 'output/Lost_GO_terms_in_five_species.xlsx')After filtering for plant-specific GO-terms
We will use the GO-terms that are plant-specific as identified by the GOMAP paper. See https://github.com/wkpalan/GOMAP-maize-analysis/blob/main/6.plantSpecific/1.getSppSpecific.R or https://plantmethods.biomedcentral.com/articles/10.1186/s13007-021-00754-1
go_plant <- read_tsv('https://raw.githubusercontent.com/wkpalan/GOMAP-maize-analysis/main/data/go/speciesSpecificGOTerms.txt')Rows: 45031 Columns: 5
── Column specification ────────────────────────────────────────────────────────
Delimiter: "\t"
chr (1): GOterm
dbl (4): NCBITaxon:10090, NCBITaxon:33090, NCBITaxon:3702, NCBITaxon:40674
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.# taxon 33090 is Viridiplantae
plantSpecificGO <- go_plant %>% dplyr::filter(`NCBITaxon:33090`==1) %>% pull(GOterm)
plantSpecificGO <- c(plantSpecificGO,c("GO:0005575","GO:0008150","GO:0003674"))results_d %>% filter(GOID %in% plantSpecificGO) %>% writexl::write_xlsx('output/Lost_GO_terms_in_five_species.PlantSpecific.xlsx')Now let’s redo the Venn diagram with those filtered GO-terms
filters <- lapply(a_go_ids, function(ch) ch %in% plantSpecificGO)
newa <- list()
for (species in names(filters)) {
before <- a[[species]]
after <- before[filters[[species]]]
newa[[species]] <- after
}plot(venn(newa),
quantities = TRUE,
fill = rev(wes_palette("Zissou1", 15, type = 'continuous')),
alpha = 0.5,
labels = list(font = 4))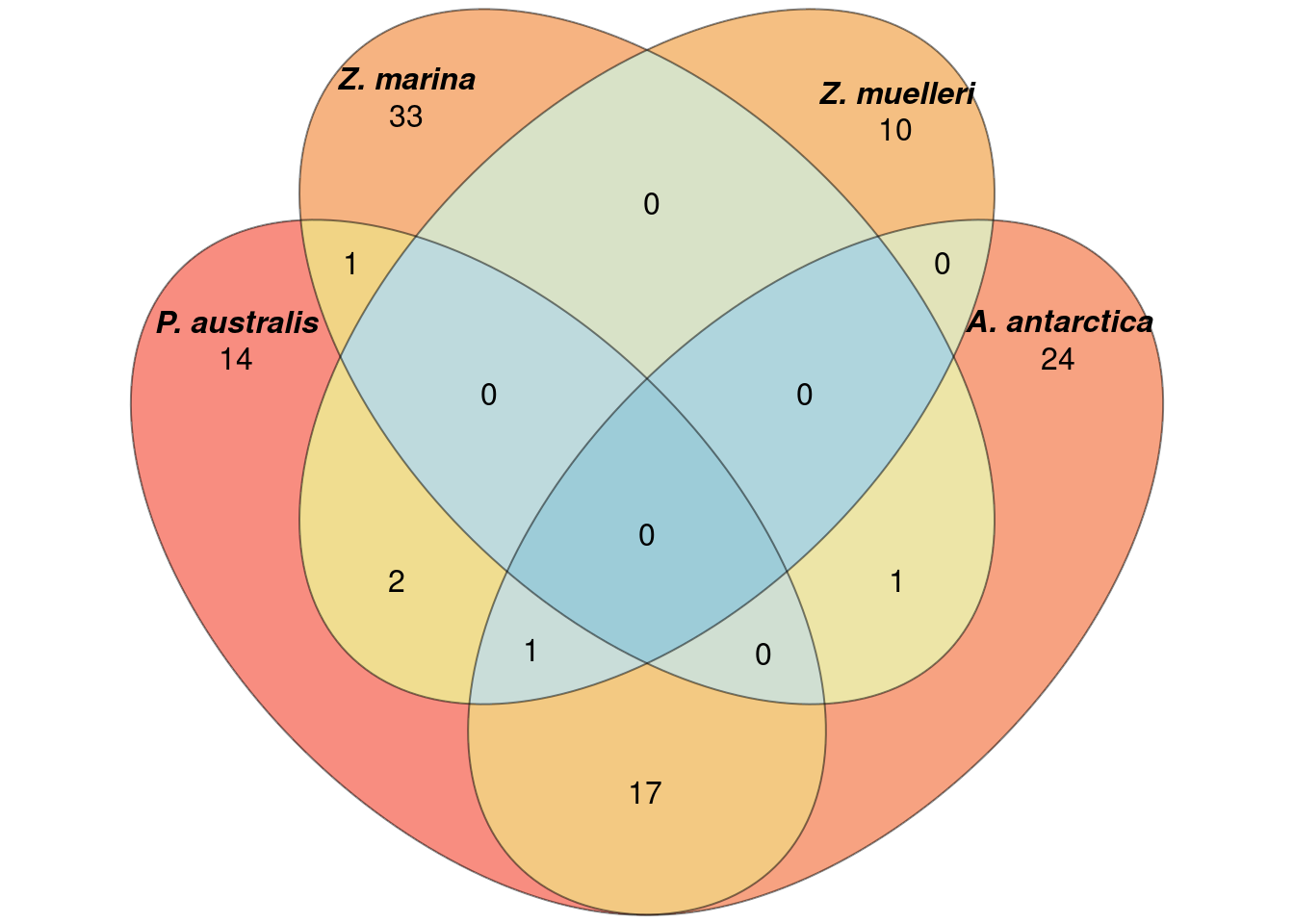
Not much difference?
sessionInfo()R version 4.3.2 (2023-10-31)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04.6 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.9.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.9.0
locale:
[1] LC_CTYPE=en_AU.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_AU.UTF-8 LC_COLLATE=en_AU.UTF-8
[5] LC_MONETARY=en_AU.UTF-8 LC_MESSAGES=en_AU.UTF-8
[7] LC_PAPER=en_AU.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_AU.UTF-8 LC_IDENTIFICATION=C
time zone: Australia/Perth
tzcode source: system (glibc)
attached base packages:
[1] stats4 stats graphics grDevices datasets utils methods
[8] base
other attached packages:
[1] patchwork_1.1.2 kableExtra_1.3.4 UpSetR_1.4.0
[4] eulerr_7.0.0 ggrepel_0.9.4 stringi_1.7.12
[7] httr_1.4.6 wesanderson_0.3.7 rrvgo_1.12.2
[10] rvest_1.0.3 lubridate_1.9.2 forcats_1.0.0
[13] stringr_1.5.0 dplyr_1.1.2 purrr_1.0.1
[16] readr_2.1.4 tidyr_1.3.0 tibble_3.2.1
[19] ggplot2_3.4.2 tidyverse_2.0.0 topGO_2.52.0
[22] SparseM_1.81 GO.db_3.17.0 AnnotationDbi_1.62.2
[25] IRanges_2.34.0 S4Vectors_0.38.1 Biobase_2.60.0
[28] graph_1.78.0 BiocGenerics_0.46.0 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] org.At.tair.db_3.17.0 RColorBrewer_1.1-3 rstudioapi_0.14
[4] jsonlite_1.8.4 umap_0.2.10.0 magrittr_2.0.3
[7] farver_2.1.1 rmarkdown_2.21 ragg_1.2.5
[10] fs_1.6.2 zlibbioc_1.46.0 vctrs_0.6.2
[13] memoise_2.0.1 RCurl_1.98-1.12 askpass_1.1
[16] webshot_0.5.5 htmltools_0.5.5 curl_5.0.0
[19] sass_0.4.6 bslib_0.4.2 plyr_1.8.8
[22] cachem_1.0.8 whisker_0.4.1 igraph_1.5.1
[25] mime_0.12 lifecycle_1.0.3 pkgconfig_2.0.3
[28] Matrix_1.6-2 R6_2.5.1 fastmap_1.1.1
[31] GenomeInfoDbData_1.2.10 shiny_1.8.0 digest_0.6.31
[34] colorspace_2.1-0 ps_1.7.5 rprojroot_2.0.3
[37] RSpectra_0.16-1 textshaping_0.3.6 RSQLite_2.3.3
[40] labeling_0.4.2 fansi_1.0.4 timechange_0.2.0
[43] polyclip_1.10-6 compiler_4.3.2 bit64_4.0.5
[46] withr_2.5.0 DBI_1.1.3 highr_0.10
[49] openssl_2.0.6 tools_4.3.2 httpuv_1.6.11
[52] glue_1.6.2 callr_3.7.3 GOSemSim_2.26.1
[55] promises_1.2.0.1 polylabelr_0.2.0 grid_4.3.2
[58] getPass_0.2-2 gridBase_0.4-7 generics_0.1.3
[61] gtable_0.3.3 tzdb_0.4.0 data.table_1.14.8
[64] hms_1.1.3 xml2_1.3.4 utf8_1.2.3
[67] XVector_0.40.0 pillar_1.9.0 vroom_1.6.3
[70] later_1.3.1 lattice_0.22-5 renv_1.0.2
[73] bit_4.0.5 tidyselect_1.2.0 tm_0.7-11
[76] Biostrings_2.68.1 knitr_1.42 git2r_0.32.0
[79] gridExtra_2.3 NLP_0.2-1 svglite_2.1.2
[82] xfun_0.39 matrixStats_1.1.0 pheatmap_1.0.12
[85] yaml_2.3.7 evaluate_0.21 wordcloud_2.6
[88] BiocManager_1.30.20 cli_3.6.1 xtable_1.8-4
[91] reticulate_1.34.0 systemfonts_1.0.4 munsell_0.5.0
[94] processx_3.8.1 jquerylib_0.1.4 treemap_2.4-4
[97] Rcpp_1.0.10 GenomeInfoDb_1.36.0 png_0.1-8
[100] parallel_4.3.2 ellipsis_0.3.2 blob_1.2.4
[103] bitops_1.0-7 viridisLite_0.4.2 slam_0.1-50
[106] scales_1.2.1 writexl_1.4.2 crayon_1.5.2
[109] rlang_1.1.1 cowplot_1.1.1 KEGGREST_1.40.1